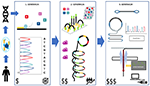
From Sanger to genome sequencing - an overview of DNA sequencing technologies
DOI:
https://doi.org/10.18388/pb.2021_534Abstract
There is no technique that would make a greater contribution to the development of genetics, molecular biology and medicine than DNA sequencing. For many years, the method based on enzymatic DNA synthesis developed by Frederic Sanger was the gold standard in this area. At the end of the 20th century, there was a dynamic development of next-generation sequencing (NGS) technologies, which ended the era of single gene analysis and initiated the era of genome sequencing. Despite fierce competition, one NGS technology has practically completely dominated the global market. In the article, we present our own review of DNA sequencing methods, starting from the Sanger method to high-throughput second- and third-generation sequencing technologies, with particular emphasis on those that have achieved commercial success. We present their short history, principles of operation, technical possibilities, applications and limitations. In the summary, we reveal how much human genome sequencing costs at the current stage of the genomic revolution and outline the prospects for further development of genomics.
Published
License
Copyright (c) 2024 Małgorzata Marcinkowska-Swojak, Magdalena Rakoczy, Jan Podkowiński, Jurand Handschuh, Paweł Wojciechowski, Luiza Handschuh

This work is licensed under a Creative Commons Attribution 4.0 International License.
All journal contents are distributed under the Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA 4.0) license. Everybody may use the content following terms: Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made, ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original. There are no additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
Copyright for all published papers © stays with the authors.
Copyright for the journal: © Polish Biochemical Society.