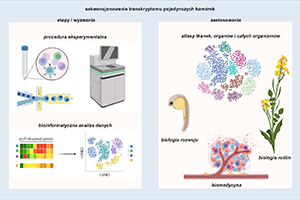
Sequencing the transcriptome of single cells: a path to discovering cellular biodiversity
DOI:
https://doi.org/10.18388/pb.2021_532Abstract
Single-cell transcriptomics (scRNA-Seq) is a breakthrough technology that has opened the way to characterizing gene expression with unprecedented resolution. It has enabled the discovery of the cellular diversity of organisms and tracing their developmental processes. A range of technological solutions have been developed to allow analysis of tens of thousands to even a million cells in a single experiment, as well as an extensive set of tools for bioinformatics analysis of the generated data. The wealth of information provided by scRNA-Seq and the possibility of using this method to study cells, organoids, tissues and even entire organisms determine its wide range of applications. In this paper, we present the experimental and computational parts of the scRNA-Seq procedure, as well as the most important applications of this technology in biomedicine, developmental biology and plant biology.
Downloads
Published
License
Copyright (c) 2024 Anna Samelak-Czajka, Małgorzata Marszałek-Zeńczak, Paulina Jackowiak

This work is licensed under a Creative Commons Attribution 4.0 International License.
All journal contents are distributed under the Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA 4.0) license. Everybody may use the content following terms: Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made, ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original. There are no additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
Copyright for all published papers © stays with the authors.
Copyright for the journal: © Polish Biochemical Society.