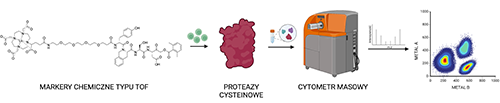
Multiparametric analysis of proteolytic enzyme activity using mass cytometry
DOI:
https://doi.org/10.18388/pb.2021_477Abstract
Proteolytic enzymes are cell catalysts that are responsible for accelerating amide bond hydrolysis in their corresponding protein substrates. Proteases are key elements in multiple cell signaling pathways and they help to maintain homeostasis. The activity of these enzymes is controlled by intracellular inhibitors that prevent excessive hydrolysis of protein substrates. Particular cases of protease gene mutations can cause serious pathophysiological conditions, such as neurodegenerative diseases, cancer or blood coagulation disorders. Therefore, there is an urgent need for a precise analysis of the activity of individual proteases. To this date, the golden standard in this type of investigation is the use of activity-based probes (ABPs) that irreversibly bind to the target protein in its catalytically active form, and enable its detection in biological samples. The main problem in dissecting protease activities inside the cell is the fact that many of them work together in extremely complex cellular signaling networks. Therefore, it is necessary to be able to perform a multi-parameter analysis that will show all of the sampled elements actively operating within a given system. This work describes the main assumptions of the doctoral dissertation of dr inż. Katarzyna Groborz, which concern the design, synthesis and validation of a new type of chemical tools for studying the activity of proteases using mass cytometry. These tools, so-called TOF probes (Time Of Flight), possess the advantages of traditional activity-based probes and also enable multi-parameter analysis of complex biological systems using high-throughput technology-mass cytometry.
Published
Issue
Section
License
Copyright (c) 2023 Katarzyna Małgorzata Groborz

This work is licensed under a Creative Commons Attribution 4.0 International License.
All journal contents are distributed under the Creative Commons Attribution-ShareAlike 4.0 International (CC BY-SA 4.0) license. Everybody may use the content following terms: Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made, ShareAlike — If you remix, transform, or build upon the material, you must distribute your contributions under the same license as the original. There are no additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
Copyright for all published papers © stays with the authors.
Copyright for the journal: © Polish Biochemical Society.